Clinical Assessment Data
AMP PD harmonizes, or standardizes, similar data collected across BioFIND, HBS, LBD, LCC, PDBP, PPMI, STEADY-PD3 and SURE-PD3. This data curation and transformation process facilitates and simplifies cross-cohort analysis. More specifically, variable names from AMP PD studies are aligned to a global mapping file and final curation is reviewed by AMP PD; this Harmonized Dictionary, based on CDISC terminology, is available as a reference for the harmonized clinical dataset and is linked with the Harmonized Assessment and Variable Matrix. Harmonized cohort data is made available in AMP PD through BigQuery.
AMP PD Quality control of the clinical data was performed by Alena Fedarovich Rancho BioSciences and Bary Landin and Dave Vismer from Technome as part of a contract with the Foundation for the National Institutes of Health (FNIH).
Harmonized Assessment & Variable Matrix
The following variables are harmonized across a breadth of standard assessments from two or more AMP PD cohorts. Click a variable to view additional details such as its definition, values, schema, and curation notes. If you want to download a version of the full AMP PD Data Dictionary, click one of the buttons below for a specific format.
To learn more, click on the Harmonized Variables.
| Assessments | Harmonized Variables | BioFIND | HBS | LBD | LCC | PDBP | PPMI | SURE-PD3 | STEADY-PD3 |
|---|---|---|---|---|---|---|---|---|---|
| Enrollment |
| ||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
| Demographics |
| ||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
| Medical History |
| ||||||||
|
| |||||||||
|
| |||||||||
| Environment Risk Factors |
| ||||||||
|
| |||||||||
|
| |||||||||
| Clinical Assessments |
| ||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
| Biospecimen Analyses |
| ||||||||
|
| |||||||||
|
| |||||||||
|
| |||||||||
|
|

Data Curation Workflow
Data from four different Parkinson’s Disease studies were harmonized to the same standard, curated and consolidated into one dataset using automated and manual approaches. To harmonize and standardize metadata for AMP PD project a global mapping file (Harmonized Dictionary) aligning variables between datasets was first created. CDISC terminology was used for harmonized variable names and descriptions when possible. A coding file was then created to decode numeric coded variables, clean-up and standardize medication names, diagnosis, level of education, etc., and align visit names between cohorts. After mapping and coding files were generated, an automated tool was applied to transform data files and perform integration of four datasets into one set of curated files. Manual inspection of transformed files followed each phase of automatic transformation. The content of each transformed file was approved by a curator and all needed adjustments were performed manually. Finally mapping files (dictionaries) for uploading data into BigQuery tables were produced by processing the content of the curated dataset using additional R-script.
Curation workflow represents three main steps
1. Data acquisition and review
2. Data harmonization
3. Data transformation/curation and QC
Data Acquisition and Review
Based on the priority assigned by the AMP PD Clinical Data Harmonization (CDH) group, the data was split into two batches: Subset 1 & Subset 2. Considerations and approach for prioritization of clinical data to be harmonized:
- Key variables critical for interpreting biological data (e.g. demographics)
- Variables to increase ease of use of biological data (e.g. genotype)
- Relevance and importance to Parkinson's disease
- Data complementary to biologic data generated through AMP PD
- Identified as the highest priority based on collective input from research experts in the PD community
Data Harmonization
![]()
Metadata variables were harmonized based on the data compatibility upon Clinical Data Harmonization (CDH) group suggestions, decisions, and final approval. CDISC terminology was used if available for Title and Description. Values of harmonized variables from different studies were standardized and included in the coding file. The coding file contains decodes for numeric coded variables, clean-up and standardize medication names, diagnosis, level of education, etc., and aligns visit names between cohorts.
Data Transformation/Curation and QC
Both automated (custom SmartConverter tool) and manual approaches were used to perform data transformations. The original data files were inspected for extended ascii characters, number of patients, visit types, codes and their decodes availability in supporting study documents. Transformation templates and coding file were prepared based on a harmonized dictionary and curation decisions to perform three rounds of transformation/consolidation using SmartConverter. After each round output files were inspected, and additional manual transformations were performed before the next round of automated transformation and after the final curation. Subset 1 and subset 2 were curated separately using the same approach described below:
Step 1: Transform Raw Data
- Prepare vocabularies and add to primary code file
- Organize data-files by study
- Create coding file and transformation template
- Run SmartConverter Round 1 and perform QC
Step 2: Transform & Consolidate
- Organize curated files into distinct study folders
- Modify transformation template
- Consolidate subset 1 and subset 2 categories
- Run SmartConverter Round 2 and perform QC
Step 3: Transform & Finalize
- Add and consolidate clinical data (e.g. missing diagnosis inputs)
- Remove and substitute fields
- Run SmartConverter Round 3 and perform QC
Clinical Data Validation Plan
The AMP PD Clinical Data Harmonization (CDH) team crafted a plan to further validate the results of the harmonization process. The purpose of the validation plan was to:
- Ensure no new errors were introduced into the clinical data as a result of the data harmonization process
- Facilitate identification of records that should be excluded from the public release
- Identify a set of tests that can be run to validate additional data submission from the current AMP PD cohorts as well as future data submissions from new cohorts
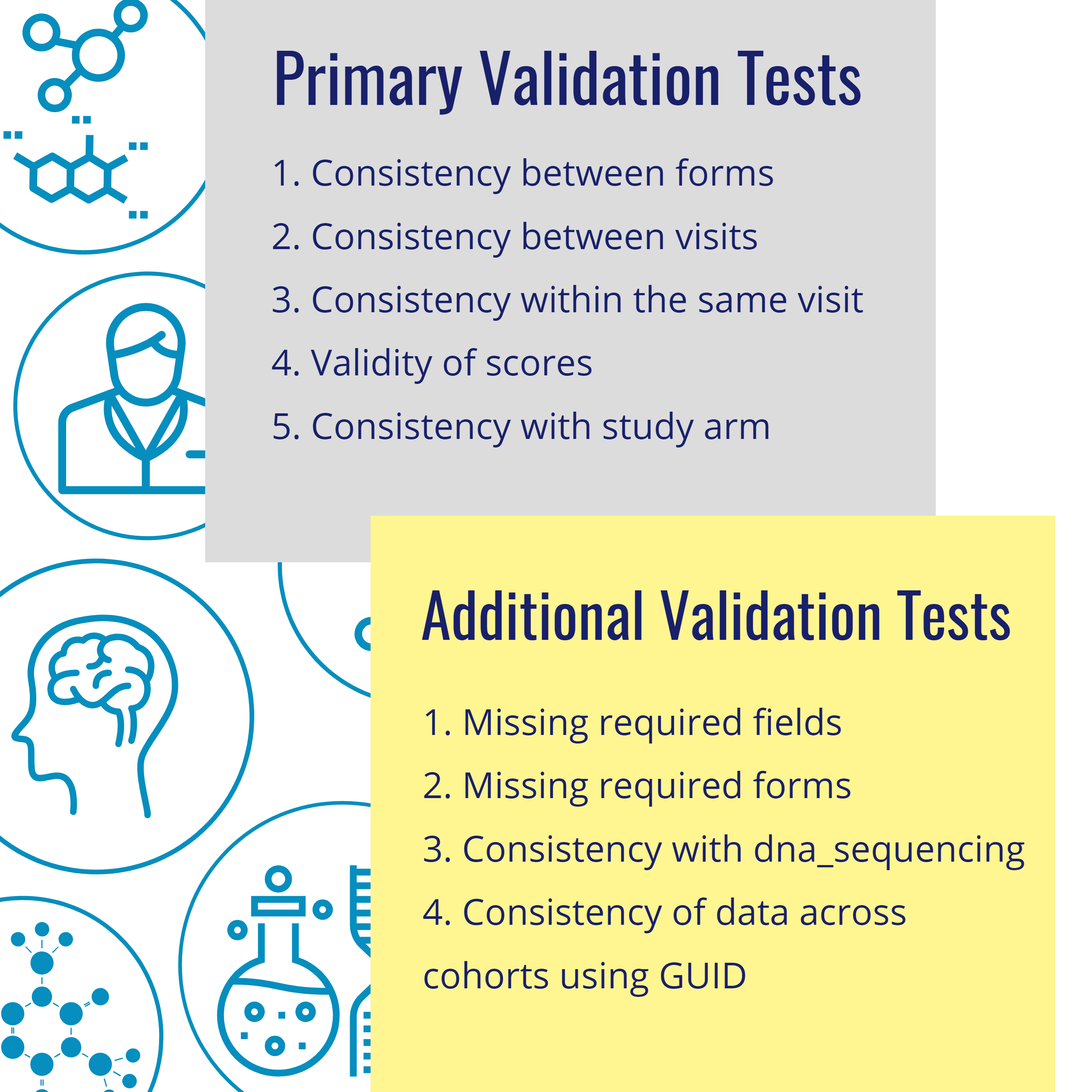 The CDH team constructed: 42 individual cohort tests, identified 23 unique tests to run against harmonized data from all four cohorts, and identified 19 tests that were not valid against harmonized data because of excluded or modified data points, or changes to data structures.
The CDH team constructed: 42 individual cohort tests, identified 23 unique tests to run against harmonized data from all four cohorts, and identified 19 tests that were not valid against harmonized data because of excluded or modified data points, or changes to data structures.
The following key decisions and outputs were made as a result of executing the validation plan:
1. Alignment of SmartConverter data outputs against program and cohort specific tests
2. Final inclusion/exclusion release criteria for clinical data
3. Secondary dataset(s) for further analysis and curation for potential future release
4. Confirmed AMP PD Subject Master List
5. Final AMP PD clinical dataset for public release
Cohort & Across Cohort Business Rules
AMP PD received cohort specific business rules from BioFIND, HBS, PDBP, and PPMI. These rules were applied by the cohorts to the raw data inputs prior to the clinical data harmonization process and succeeding datasets were required to follow these business rules. As part of the QC process, these business rules were re-checked after the harmonization process to ensure the rules were still valid.