News & Updates
AMP PD Release Notes – December 2020
Data Summary
Data Composition
Clinical Data
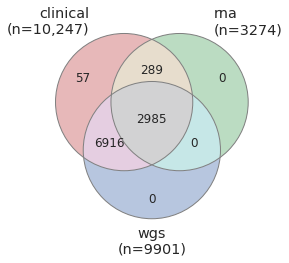 Participant records were compiled from seven cohorts and harmonized to form a single unified AMP PD cohort dataset. These records were then paired with RNA and WGS samples and excluded if matching sample data was not available, with the exception of 57 participants who appeared in multiple studies and whose duplicate WGS samples were excluded. Participants appearing in multiple studies and their corresponding samples are identified in release products so they may be traced to their associated clinical records from multiple studies. Growing from 4298 participants in our flagship launch on October 15, 2019, now 10,247 participants are represented in this v2 release by clinical records and at least one other data type.
Participant records were compiled from seven cohorts and harmonized to form a single unified AMP PD cohort dataset. These records were then paired with RNA and WGS samples and excluded if matching sample data was not available, with the exception of 57 participants who appeared in multiple studies and whose duplicate WGS samples were excluded. Participants appearing in multiple studies and their corresponding samples are identified in release products so they may be traced to their associated clinical records from multiple studies. Growing from 4298 participants in our flagship launch on October 15, 2019, now 10,247 participants are represented in this v2 release by clinical records and at least one other data type.
Integrated Data
This release includes 2985 subjects with fully integrated clinical records, WGS samples, and RNA samples. For an additional 289 participants, this release includes RNA samples with corresponding clinical records where WGS is not available. There are similarly 6916 WGS samples with clinical records where RNA sample data is not available.
RNA Data
RNA sample data was sequenced and processed for BioFIND, PDBP, and PPMI cohort participants. The AMP PD v1 release featured 8356 RNA samples for 3225 Participants. In the latest v2 release, 105 RNA samples have been added, bringing the total participants in AMP PD with RNA samples to 3274. RNA samples were excluded from the v2 release when there was no corresponding clinical data. All RNA samples were vetted through a series of independent genomic QC checks and interdependent multi-modal QC checks.
WGS Data
DNA samples were sequenced and processed through the Broads GATK pipeline for BioFIND, HBS, LBD, LCC, PDBP, PPMI, and Steady PD cohort participants. WGS samples were excluded from the v2 release when there was no corresponding clinical data. All WGS samples were vetted through a series of independent genomic QC checks and interdependent multi-modal QC checks. In Q3 2020, AMP PD added TOPMed joint genotyped bcf data for 4047 AMP PD participants. In the v2 release dataset, all 9887 WGS samples are represented in the AMP PD Broad joint discovery vcf data, which excludes 14 released samples that are flagged for further investigation. QC flags are identified and described in AMP PD release products for each WGS sample in the v2 release.
Composition by Cohort
BioFIND Data
Participants from the BioFIND cohort are represented in AMP PD clinical, RNA, and WGS data. Of 213 participants whose clinical records met AMP PD minimum clinical data criteria, 172 have corresponding WGS sample data (3 are represented by a linked WGS duplicate sample), 172 in the AMP PD joint genotyping dataset, 208 have corresponding RNA sample data, and 167 participants have corresponding samples in all three release data categories.
HBS Data
Participants from the Harvard Biomarkers Study (HBS) are represented in AMP PD clinical and WGS data. Of 1189 HBS participants whose clinical records met AMP PD minimum clinical data criteria, 1180 have corresponding WGS sample data (9 are represented by a linked WGS duplicate sample) and 1173 are represented in the AMP PD joint genotyping dataset.
PDBP Data
Participants from the Parkinson’s Disease Biomarkers Program (PDBP) are represented in AMP PD clinical, RNA, and WGS data. Of 1606 participants whose clinical records met AMP PD minimum clinical data criteria, 1505 have corresponding WGS sample data (7 are represented by a linked WGS duplicate sample), 1500 in the AMP PD joint genotyping dataset, 1484 have corresponding RNA sample data, and 1380 participants have corresponding samples in all three release data categories.
PPMI Data
Participants from the Parkinson’s Progression Markers Initiative (PPMI) are represented in AMP PD clinical, RNA, and WGS data. Of 1923 participants whose clinical records met AMP PD minimum clinical data criteria, 1775 have corresponding WGS sample data (6 are represented by a linked WGS duplicate sample), 1773 in the AMP PD joint genotyping dataset, 1582 have corresponding RNA sample data, and 1433 participants have corresponding samples in all three release data categories.
LBD Data
Participants from the Lewy Bodies Dementia (LBD) cohort are represented in AMP PD clinical and WGS data. Of 4586 LBD participants whose clinical records met AMP PD minimum clinical data criteria, 4579 have corresponding WGS sample data (7 are represented by a linked WGS duplicate sample) and 4579 are represented in the AMP PD joint genotyping dataset.
LCC Data
Participants from the LRRK2 Cohort Consortium (LCC) cohort are represented in AMP PD clinical and WGS data. Of 638 LCC participants whose clinical records met AMP PD minimum clinical data criteria, 599 have corresponding WGS sample data (39 are represented by a linked WGS duplicate sample) and 599 are represented in the AMP PD joint genotyping dataset.
Steady-PD
Participants from the Steady-PD cohort are represented in AMP PD clinical and WGS data. Of 92 Steady-PD cohort participants whose clinical records met AMP PD minimum clinical data criteria, 91 have corresponding WGS sample data (1 is represented by a linked WGS duplicate sample) and 91 are represented in the AMP PD joint genotyping dataset.
V1 Release vs V2 Release Summary
Additions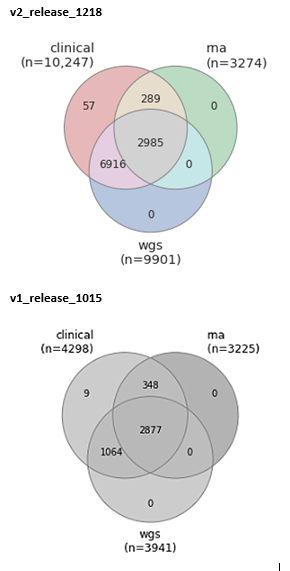
- Added a new cohort: LBD clinical and WGS samples
- Added a new cohort: LCC clinical and WGS samples
- Added a new cohort: Steady clinical and WGS samples
- Added to HBS, PDBP, and PPMI cohorts: clinical, WGS samples, and RNA samples
- Added all QC passing samples to AMP PD joint genotyping
- Added TOPMed Joint call bcf files
- Added Plink 1.9 and 2.0 data
Changes
- Modified WGS QC Process to resolve heterozygosity skew
- Flagged v1_release samples that failed v2_release QC
- Added Flags field to wgs_sample_inventory table
- Added Flag Descriptions Table
- Modified the mutations table, excluding SNCA variant H50Q (rs201106962) and changed the names on the names on the website to traditional numbering in lieu of amino acid change
- Replaced and reformatted the duplicate_participants table
- Discontinued gatk_all_variants Table