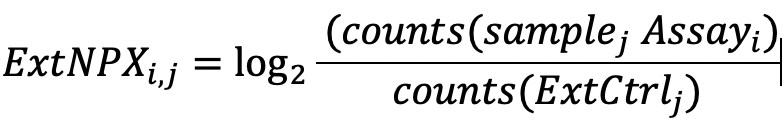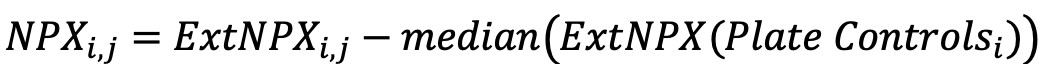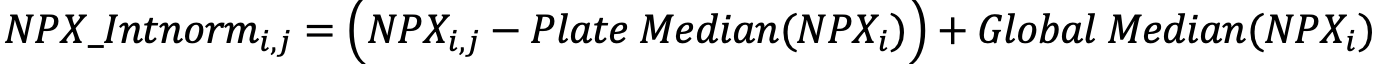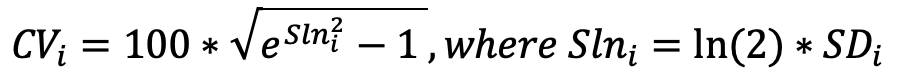Targeted Proteomics Data
AMP PD features two separate Targeted Proteomics datasets and one Targeted Proteomics release product. AMP PD Release 4.0 contains a bridged data release product generated from Release 3.0 datasets, containing samples from the NINDS Parkinson’s Disease Biomarkers Program (PDBP) and Michael J Fox Foundation’s Parkinson’s Progression Markers Initiative (PPMI).
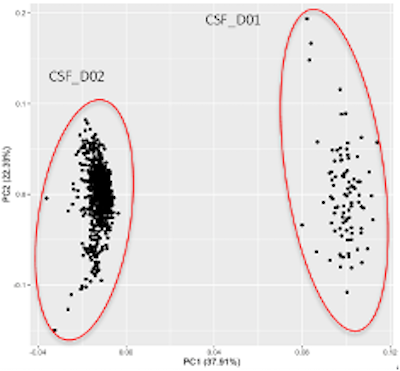
Release 3.0 contains two datasets generated using Olink 1536 from two separate data providers, labeled ‘D01’ and ‘D02’. Within the dataset ‘D01’, there were 746 CSF and Plasma samples from 213 participants. Dataset ‘D02’ includes 666 CSF samples & 898 plasma samples from 225 participants matched across both tissue types. This results in a total of 3050 samples from 413 participants.
These Targeted Proteomics datasets and release product contain eight unfiltered NPX files from four separate Panels for both Plasma and CSF samples. The four targeted proteomics panels are Cardiometabolic, Inflammation, Neurology, and Oncology.
Release 4.0 contains a bridged data release product that was generated using Olink provided bridging scripts in R. This is available under the Release 4.0 tag within the proteomics folder. In Release 3.0, all datasets are listed under the main datasets tab within the proteomics folder. All data products are split into four separate datasets. The data is split by tissue source (CSF, Plasma), data set number (D01, D02) and release (D03). From there, the main folders for each data set contain data from all four panels in both long and matrix format. An additional format is provided in the olink-explore-format folder. This format is provided for those who wish to use Olink specific tools. Each dataset also contains a sample metadata sheet for user reference.
Data has been reformatted from the original format to contain AMP PD specific participant and sample IDs. Additional QC columns have been added to the data in order to provide additional information for flagged or passed samples. The released data has accompanying Terra notebooks, ranging from “Getting Started” notebooks which help users get familiar with the data and eventually assign case and control to samples, to QC notebooks to show users QC criteria used in flagging samples and analysis notebooks to perform simple data visualizations.
As AMP PD accumulates datasets of the same source tissue and compatible methods, updates to the normalized aggregate release product will be made. Datasets in the datasets directory should not be combined in researchers' analyses without first assessing compatibility and normalizing their values, therefore, users wishing to use NPX values from both ‘D01’ and ‘D02’ datasets should use the Release 4.0 Proteomics Release product.
What's on this page:
| Baseline | 3M | 6M | 9M | 12M | 18M | 24M | 30M | 36M | 42M | 48M | 54M | 60M | 72M | 84M | 96M | Total | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDBP | Plasma | 111 | 0 | 2 | 0 | 91 | 28 | 114 | 0 | 110 | 0 | 48 | 0 | 17 | 0 | 0 | 0 | 521 |
| CSF | 111 | 0 | 2 | 0 | 91 | 28 | 114 | 0 | 110 | 0 | 48 | 0 | 17 | 0 | 0 | 0 | 521 | |
| PPMI | Plasma | 263 | 26 | 125 | 9 | 128 | 24 | 166 | 26 | 94 | 18 | 148 | 15 | 55 | 7 | 5 | 1 | 1120 |
| CSF | 244 | 5 | 111 | 0 | 109 | 0 | 152 | 1 | 84 | 0 | 133 | 1 | 43 | 0 | 5 | 1 | 888 |
Sample Selection Criteria
The criteria used to select these samples were as follows:
- Samples for three timepoint or more available
- Participant samples selected were from participants who had previously generated corresponding Whole Genome Sequencing or Transcriptomic data on the AMP PD Knowledge Platform
- All CSF samples selected had hemoglobin < 100 ng/mL to assure limited blood contamination
- All Plasma and CSF samples were collected under similar protocols
AMP PD Quality control of the preview release data was performed by Victoria Dardov from Technome as part of a contract with the Foundation for the National Institutes of Health (FNIH).
Information here was prepared by the Olink Proteomics in consultation with the AMP PD Proteomics Working Group.
Method
{"preview_thumbnail":"/sites/default/files/styles/video_embed_wysiwyg_preview/public/video_thumbnails/UFvPNPyueNc.jpg?itok=NqCC4IQa","video_url":"https://youtu.be/UFvPNPyueNc","settings":{"responsive":1,"width":"854","height":"480","autoplay":1},"settings_summary":["Embedded Video (Responsive, autoplaying)."]}
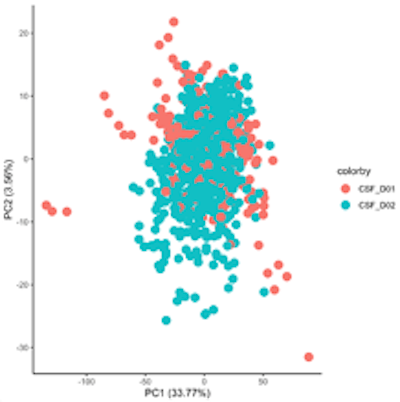
Proximity Extension Assay for Targeted Proteomics
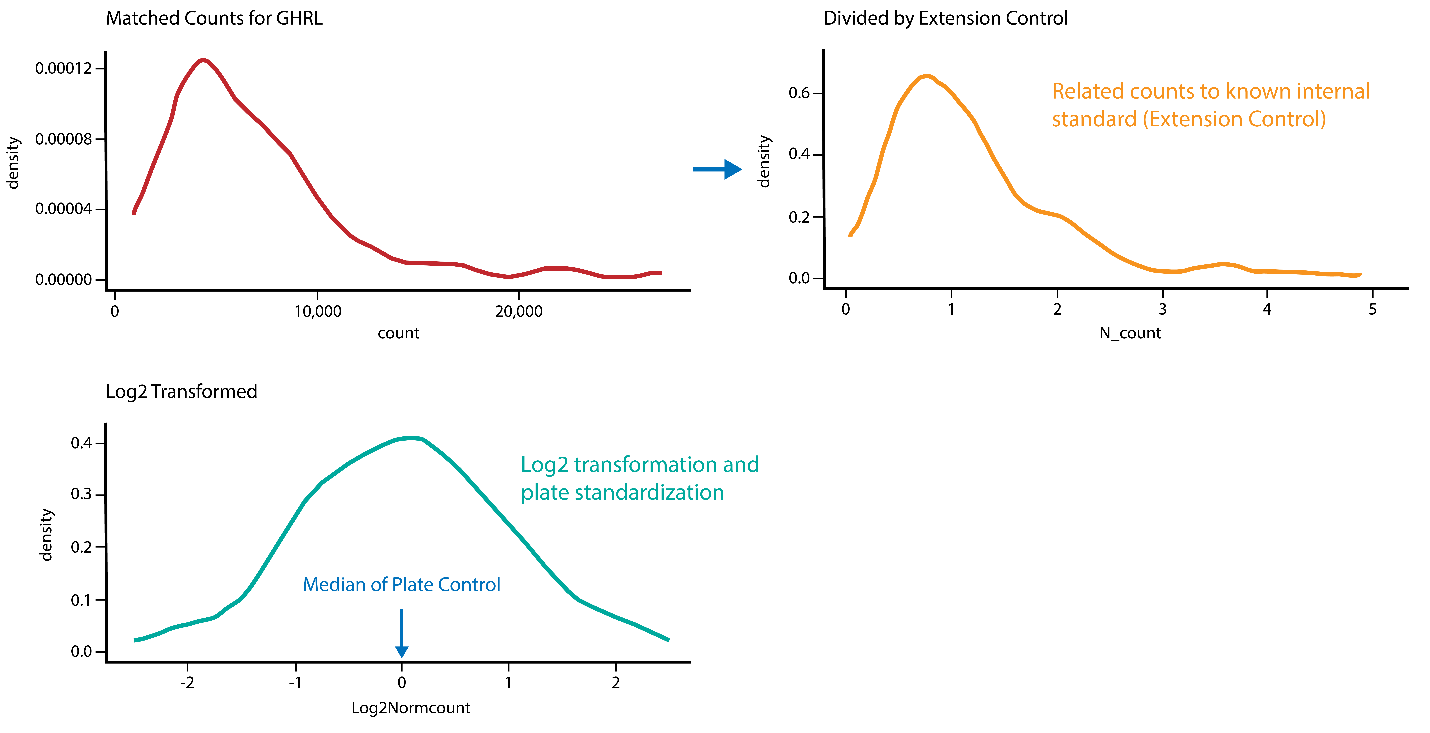
Normalized Protein Expression (NPX) quantifies the relative amount of a specific protein. This is determined by performing an immunoassay for a targeted protein. Antibodies that bind to a protein of interest contain unique sequences that are extended, amplified and subsequently detected and quantified by NGS. The amount of this sequence is normalized to standard plate controls to give relative quantities of targeted proteins.
Assay Controls Summary
Extensive quality control is performed for each assay in order to control and assess technical performance of the assay at each step. This ensures generation of reliable data.
AMP PD Controls
AMP-PD quality control further examines the data and includes sample, run and control sample QC.
Generating NPX values
The Explore system´s raw data output are NGS counts, where each combination of an assay and sample is given an integer value based on the number of DNA copies detected. These raw data counts are converted into NPX values for use in downstream statistical analysis.